Pseudomonas aeruginosa PAO1, PA3554 (arnA)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
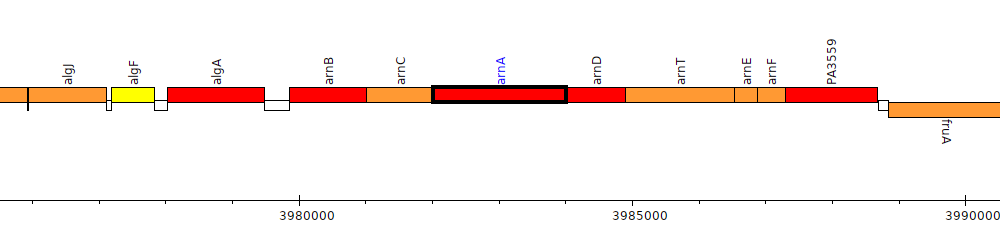
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA3554
|
| Name |
arnA
Synonym: arnA |
| Replicon | chromosome |
| Genomic location | 3982021 - 3984009 (+ strand) |
| Transposon Mutants | 3 transposon mutants in PAO1 |
| Transposon Mutants in orthologs | 2 transposon mutants in orthologs |
Cross-References
| RefSeq | NP_252244.1 |
| GI | 15598750 |
| Affymetrix | PA3554_at |
| Entrez | 878473 |
| GenBank | AAG06942.1 |
| INSDC | AAG06942.1 |
| NCBI Locus Tag | PA3554 |
| protein_id(GenBank) | gb|AAG06942.1|AE004776_5|gnl|PseudoCAP|PA3554 |
| TIGR | NTL03PA03554 |
| UniParc | UPI00000C5A65 |
| UniProtKB Acc | Q9HY63 |
| UniProtKB ID | ARNA_PSEAE |
| UniRef100 | UniRef100_Q9HY63 |
| UniRef50 | UniRef50_O52325 |
| UniRef90 | UniRef90_Q9HY63 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
ArnA
|
| Synonyms |
PmrI ArnA |
| Evidence for Translation |
Identified using nanoflow high-pressure liquid chromatography (HPLC) in conjunction with microelectrospray ionization on LTQ XL mass spectrometer (PMID:24291602).
Detected using LC-MS/MS (74.36 kDa, PMID:21751344)
|
| Charge (pH 7) | -3.52 |
| Kyte-Doolittle Hydrophobicity Value | -0.253 |
| Molecular Weight (kDa) | 74.4 |
| Isoelectric Point (pI) | 6.68 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
AMR gene predictions from CARD database
Identified using the Resistance Gene Identifier (RGI)
| Model Name | Definition | Accession | Model Type (?) | SNP | Cutoff | Bit Score | Percent Identity | CARD Version |
| arnA | arnA modifies lipid A with 4-amino-4-deoxy-L-arabinose (Ara4N) which allows gram-negative bacteria to resist the antimicrobial activity of cationic antimicrobial peptides and antibiotics such as polymyxin. arnA is found in E. coli and P. aeruginosa. [PMID:11021929, PMID:15939024] | ARO:3002985 | protein homolog model | Perfect | 1363.2 | 100.0 | 3.2.5 |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 651 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG002112 (427 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA3554
Search term: arnA
Search term: ArnA
|
Human Homologs
|
Ensembl
110, assembly
GRCh38.p14
|
UDP-glucuronate decarboxylase 1 [Source:HGNC Symbol;Acc:HGNC:17729]
E-value:
1.5e-27
Percent Identity:
27.3
|
|
Ensembl
110, assembly
GRCh38.p14
|
UDP-glucuronate decarboxylase 1 [Source:HGNC Symbol;Acc:HGNC:17729]
E-value:
1.5e-27
Percent Identity:
27.3
|
References
|
PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance.
Gunn JS, Lim KB, Krueger J, Kim K, Guo L, Hackett M, Miller SI
Mol Microbiol 1998 Mar;27(6):1171-82
PubMed ID: 9570402
|
|
A formyltransferase required for polymyxin resistance in Escherichia coli and the modification of lipid A with 4-Amino-4-deoxy-L-arabinose. Identification and function oF UDP-4-deoxy-4-formamido-L-arabinose.
Breazeale SD, Ribeiro AA, McClerren AL, Raetz CR
J Biol Chem 2005 Apr 8;280(14):14154-67
PubMed ID: 15695810
|
|
Structure and function of both domains of ArnA, a dual function decarboxylase and a formyltransferase, involved in 4-amino-4-deoxy-L-arabinose biosynthesis.
Williams GJ, Breazeale SD, Raetz CR, Naismith JH
J Biol Chem 2005 Jun 17;280(24):23000-8
PubMed ID: 15809294
|
|
Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa.
Choi DS, Kim DK, Choi SJ, Lee J, Choi JP, Rho S, Park SH, Kim YK, Hwang D, Gho YS
Proteomics 2011 Aug;11(16):3424-9
PubMed ID: 21751344
|